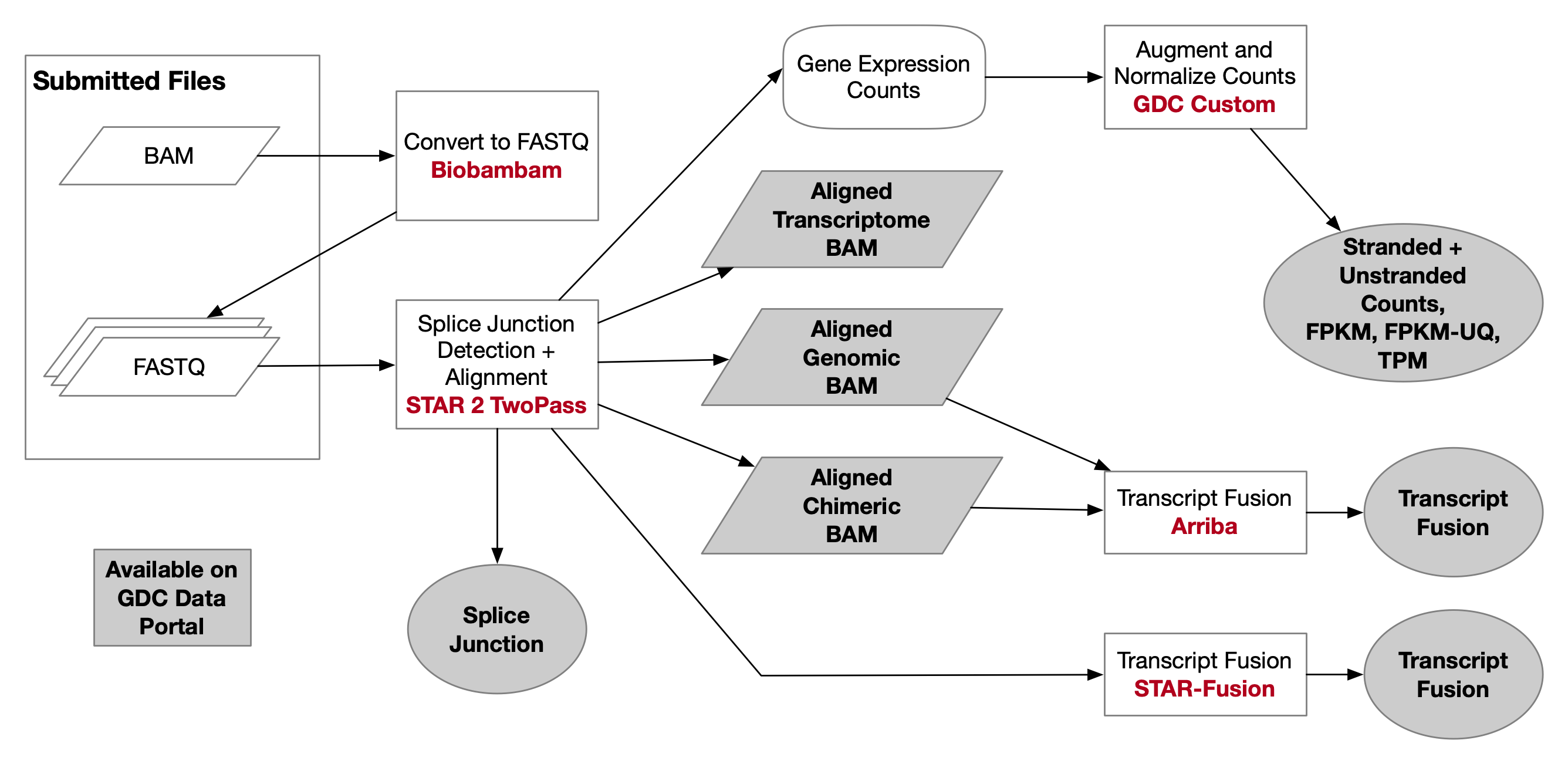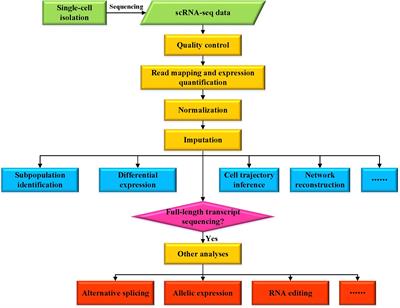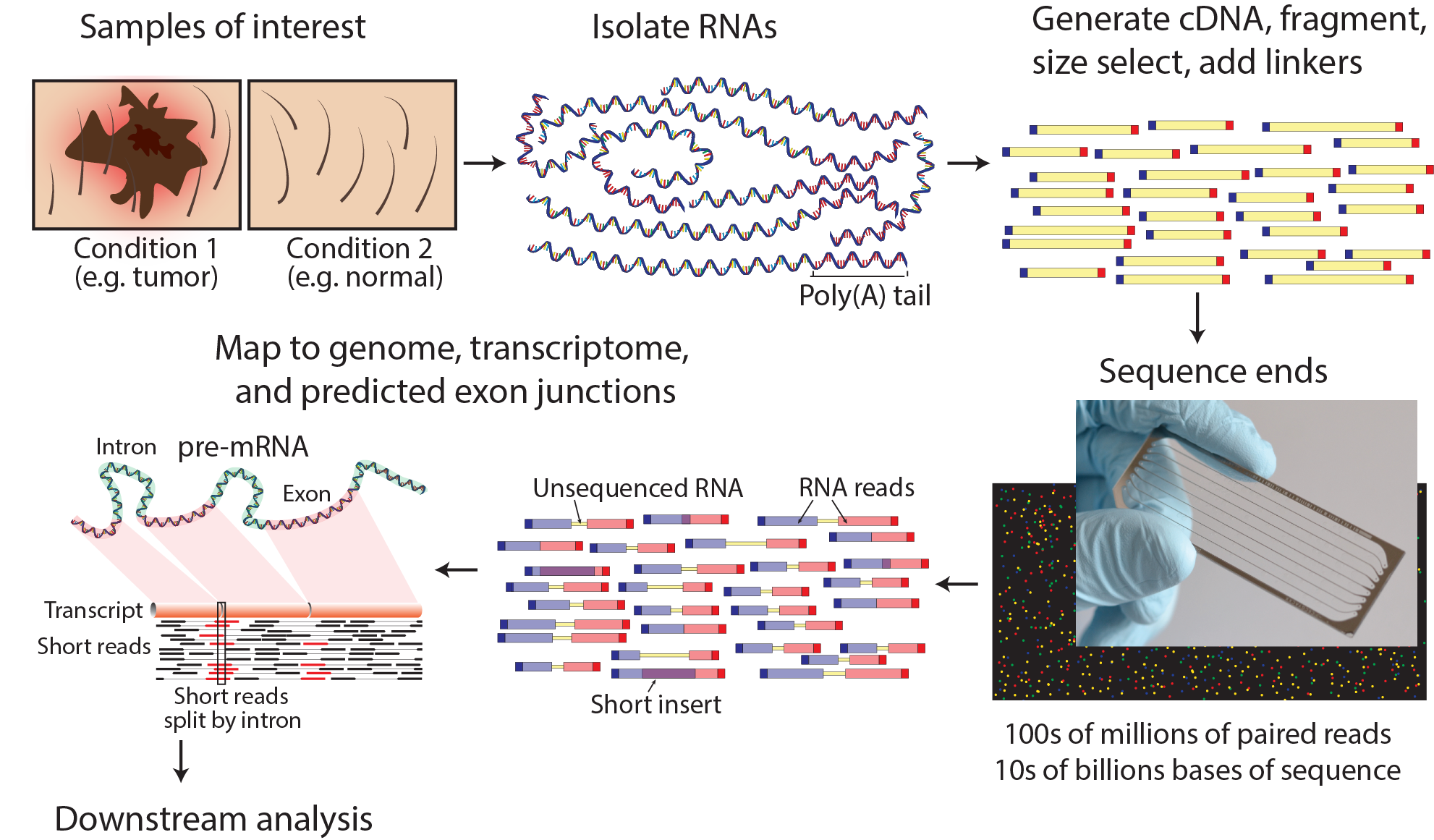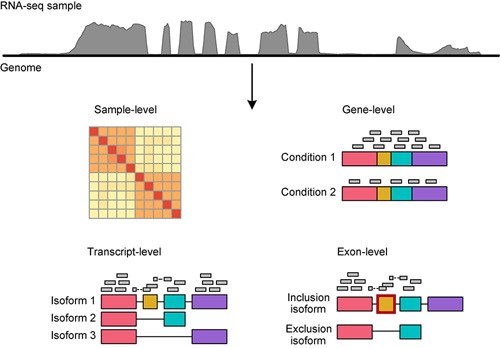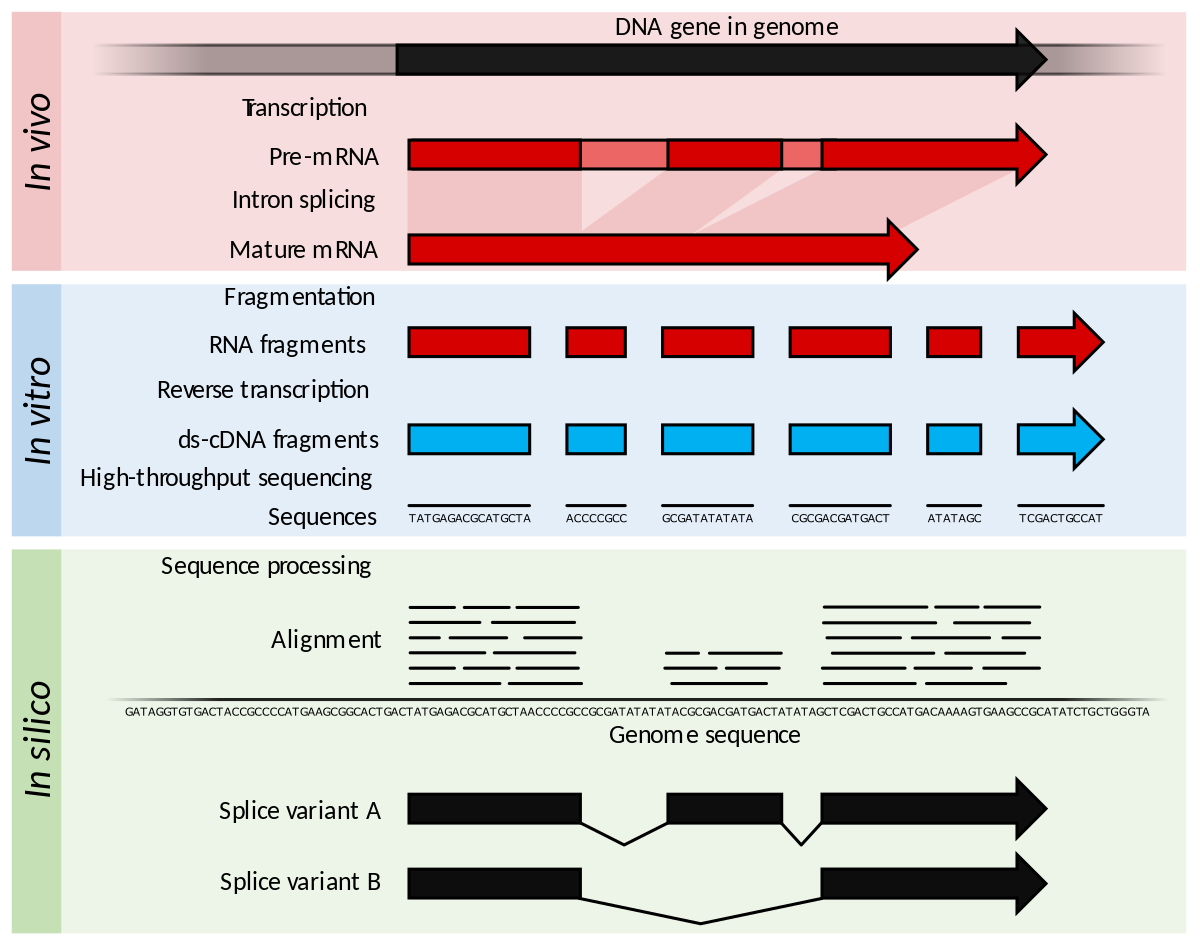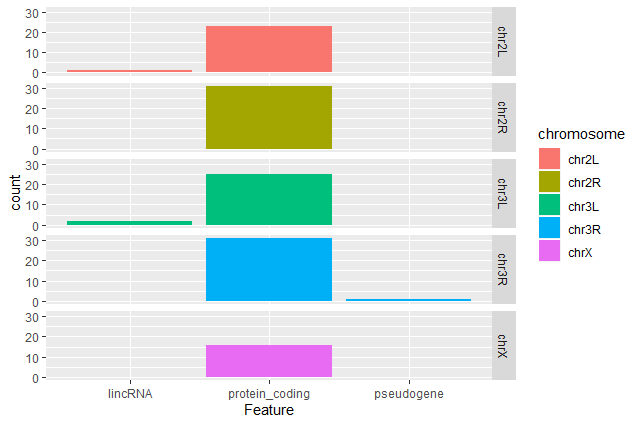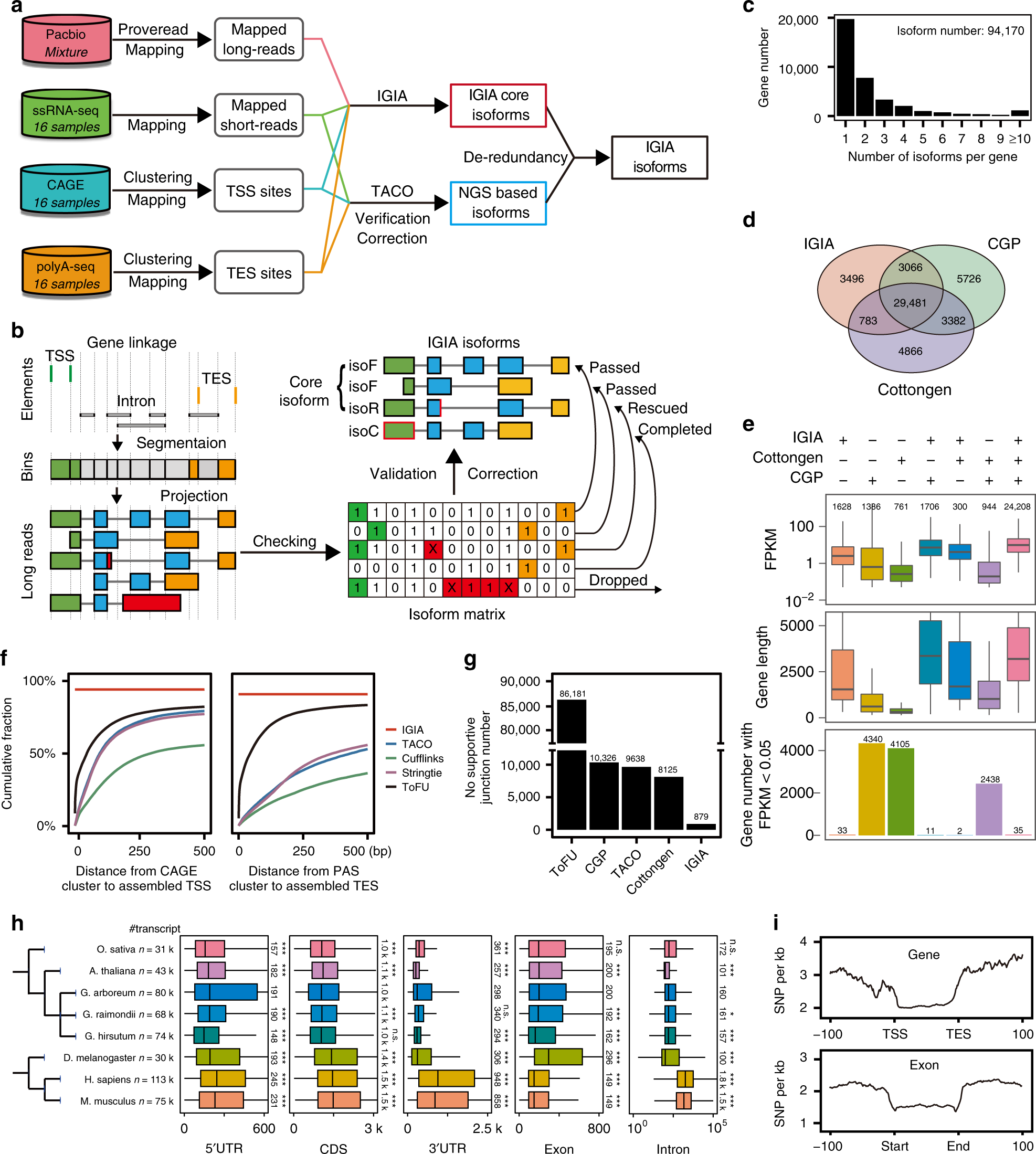
Multi-strategic RNA-seq analysis reveals a high-resolution transcriptional landscape in cotton | Nature Communications
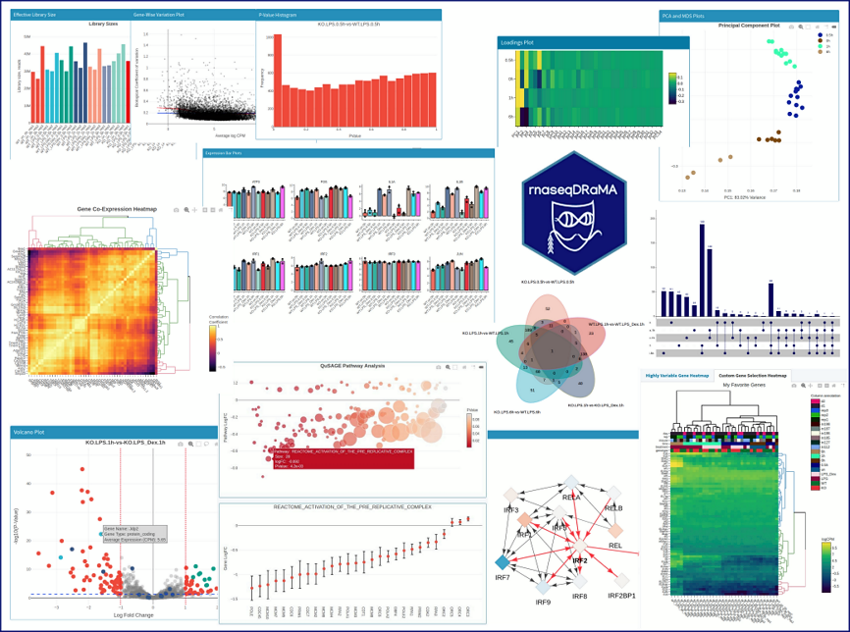
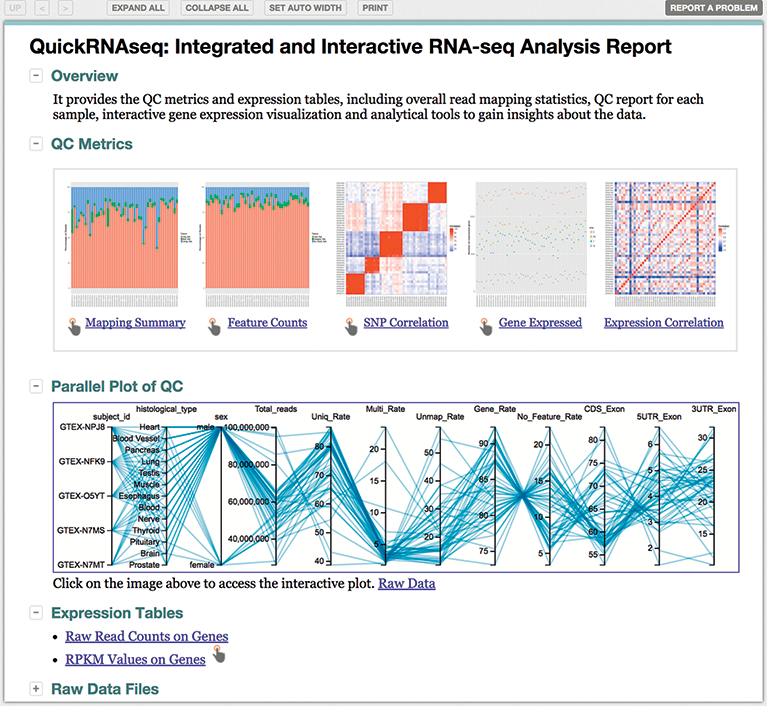
rnaseqDRaMA - RNAseq data visualization and mining - 2020 Shiny Contest Submission - Shiny Contest - Posit Community
![PDF] CellBench: R/Bioconductor software for comparing single-cell RNA-seq analysis methods | Semantic Scholar PDF] CellBench: R/Bioconductor software for comparing single-cell RNA-seq analysis methods | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/25de217a392177051982f995cd276300ab3a13fa/2-Figure1-1.png)
PDF] CellBench: R/Bioconductor software for comparing single-cell RNA-seq analysis methods | Semantic Scholar

Protocol for analysis of RNA-sequencing and proteome profiling data for subgroup identification and comparison - ScienceDirect
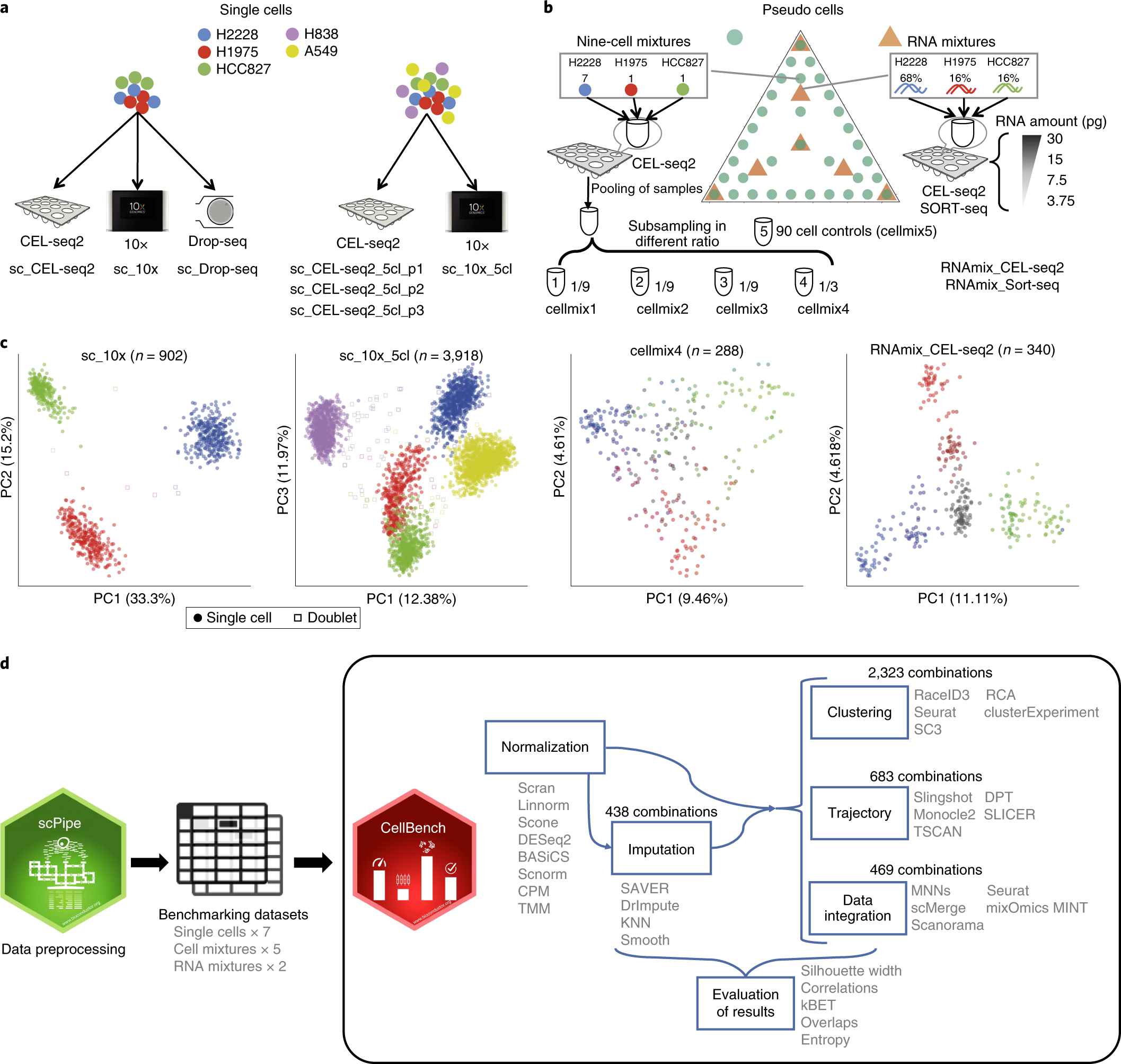
Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments | Nature Methods
![Hands-on: Single-Cell RNA-Sequencing Data Analysis Using Command-Line and R [Complete Training] - BioCode Hands-on: Single-Cell RNA-Sequencing Data Analysis Using Command-Line and R [Complete Training] - BioCode](https://i0.wp.com/www.biocode.org.uk/wp-content/uploads/2023/02/Course-Thumbnail-for-Hands-on-Single-Cell-RNA-Sequencing-Data-Analysis-Using-Command-Line-and-R.webp?fit=1920%2C1080&ssl=1)